
Description
Welcome to the iLund4u phage database!
iLund4u is a bioinformatics tool designed for the search and annotation of accessory genes and hotspots across large datasets. We recommend visiting the tool's Home page, where you can find a detailed description of the algorithm and full documentation.
This iLund4u phage database provides an interactive platform to explore hotspot-encoded phage proteins annotated with iLund4u, using the complete PhageScope database of 873K phage genomes as input.

You can also access the web version of iLund4u on our server portal which can be used to find whether homologues of your query protein are encoded in hotspots.
The results generated in the web version are directly linked to the data and pages of this database.
Database overview
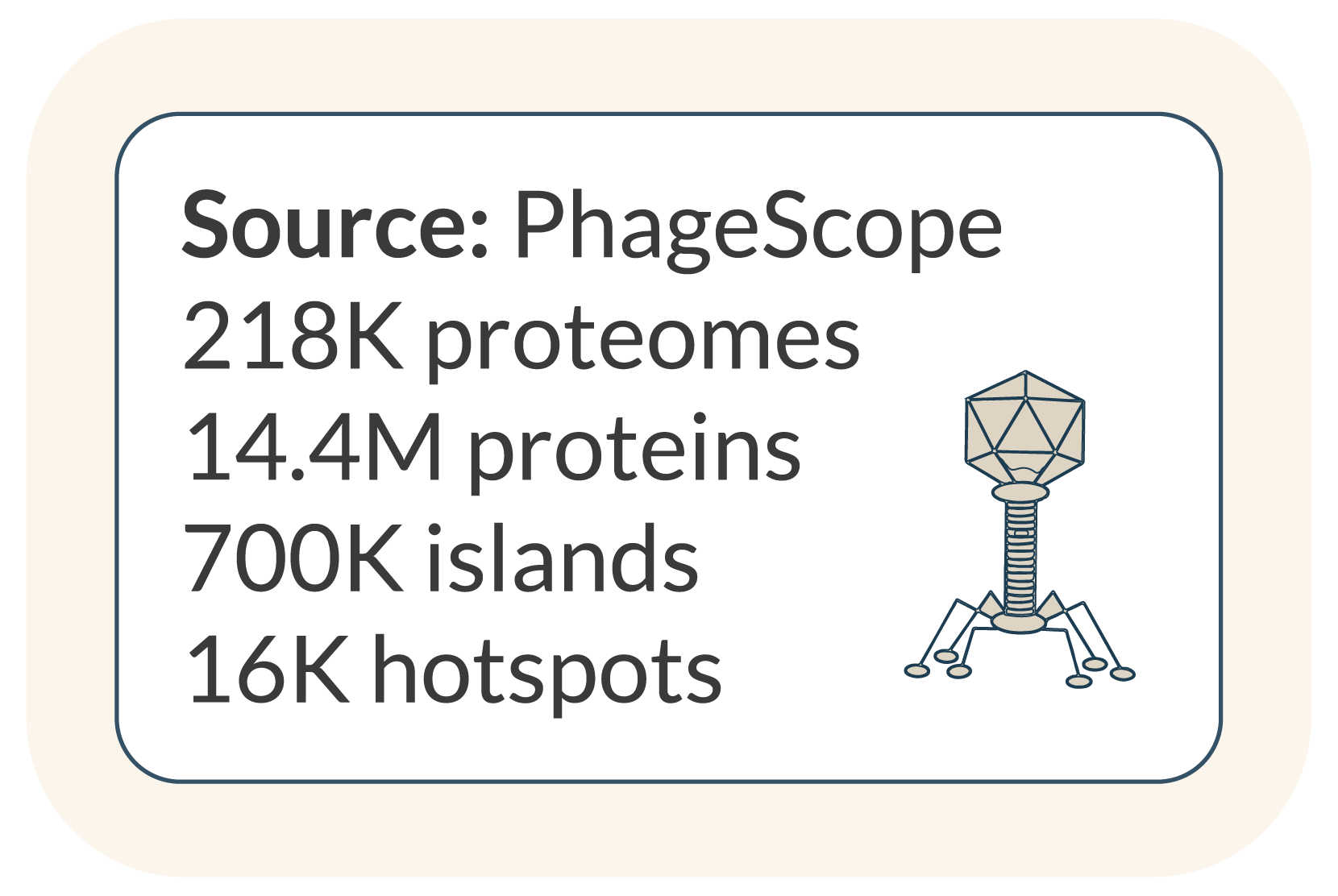
iLund4u includes two precomputed hotspot databases derived from phage and plasmid sequences. This online resource catalogues phage-associated hotspots and accessory protein families. It consists of 242.8K protein families encoded in 16K hotspots across 218K phage genomes, remaining after deduplication and filtering of the initial PhageScope dataset.
Protein Families
Here you can find all protein families encoded at least once as cargo in phage hotspots — a total of 242,773 representative sequences. Each protein family has its own database page with a detailed description. Note that you can also find families homologous to your query protein using the iLund4u web search tool.
Phages
This dataset includes 217,963 phage genomes that remained after deduplication and filtering of the initial PhageScope collection using the iLund4u algorithm. Each phage has its own database page containing description and classification of its proteins as variable, conserved, or intermediate. In addition, each phage is assigned to a proteome community — a group of related phages sharing a core proteins. You can also explore proteome communities and identify phages related to your query genome using the iLund4u web search tool.
Reference
If you find iLund4u useful, please cite:
Artyom. A. Egorov, Vasili Hauryliuk, Gemma C. Atkinson, Systematic
annotation of hyper-variability hotspots in phage genomes and plasmids
bioRxiv 22024.10.15.618418; doi:
10.1101/2024.10.15.618418
Authors & Contact
iLund4u is developed by Artyom Egorov at the Atkinson Lab, Department of
Experimental Medical Science, Lund University, Sweden.
We are open for suggestions of how we can extend and improve iLund4u functionality.
Please
don't hesitate to share your ideas or
feature requests.
Please contact us by e-mail or use GitHub Issues to report any technical problems related to iLund4u.